JCRB1030 JHH-6
Cell information
Important Notice(s)On the agreement for distribution of JHH series cell lines, NOZ and OZ
Cell type:general cells (View Pricing Information)
| JCRB No. | JCRB1030 | Cell Name | JHH-6 |
|---|---|---|---|
| Profile | Human hepatocellular carcinoma cell line. | Other Name | |
| Animal | human | Strain | |
| Genus | Homo | Species | sapiens |
| Sex | F | Age | 57 |
| Identity | available | Tissue for Primary Cancer | liver, gallbladder |
| Case history | anhepatic liver carcinoma, hetocellular carcinoma (Ed.II). HBs-Ag(-), HBs-Ab(-), HBe-Ag(-), HBc-Ab(-), AFP(ND), CEA 4.6ng/ml. | Metastasis | |
| Tissue Metastasized | Genetics | HBV-DNA was not integrated | |
| Life Span | infinite | Crisis PDL | |
| Morphology | ephitherial-like structure | Character | Undifferentiated morphology observed. Albumin, AFP and CEA were not observed in the supernatant of the cell culture but ferritin was observed (6.7 ng/ml). HBs-Ag(-), HBs-Ag(-). |
| Classify | tumor | Established by | Matsuura,T. & Nagamori,S. |
| Registered by | Nagamori,S. | Regulation for Distribution | Require negotiation and agreement from Dr.Nagamori before shipping. |
| Comment | Cell Bank will handle to get the approval but the depositor may contact to the requested person directly. | Year | 2001 |
| Medium | Williams'E medium with 10% fetal calf serum. | Methods for Passages | Cells are harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Cell Number on Passage | not defined | Race | Japanese |
| CO2 Conc. | 5 % | Tissue Sampling | |
| Tissue Type | undifferentiated hepato cellular carcinoma |
| Detection of virus genome fragment by Real-time PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Detected DNA Virus | tested | Detected RNA Virus | tested | ||||||
| CMV | - | parvoB19 | - | HCV | - | HTLV-1 | - | ||
| EBV | - | HBV | - | HIV-1 | - | HTLV-2 | - | ||
| HHV6 | - | HTLV-1 | - | HIV-2 | - | HAV | - | ||
| HHV7 | - | HTLV-2 | - |
-/negative. +/positive. nt/not tested. (positive (+) does not immediately mean the production of infectious viral particles.) |
|||||
| BKV | - | HIV-1 | - | ||||||
| JCV | - | HIV-2 | - | ||||||
| ADV | - | HPV18 | - | ||||||
| Notes | |||||||||
| Reference | |
|---|---|
| Pubmed id:1701409 | Integration of hepatitis B virus DNA into cells of six established human hepatocellular carcinoma cell lines. Fujise K,Nagamori S,Hasumura S,Homma S,Sujino H,Matsuura T,Shimizu K,Niiya M,Kameda H,Fujita K Hepatogastroenterology. 1990 Oct;37(5):457-60 |
| Pubmed id:2484807 | [Protein secretion of human cultured liver cells]. Nagamori S,Fujise K,Hasumura S,Homma S,Sujino H,Matsuura T,Shimizu K,Niiya M,Kameda H Hum Cell. 1988 Dec;1(4):382-90 |
| Research results by users. Click! | |
|---|---|
| Pubmed id:24574752 |
Bortezomib effect on E2F and cyclin family members in human hepatocellular carcinoma cell lines. Baiz D,Dapas B,Farra R,Scaggiante B,Pozzato G,Zanconati F,Fiotti N,Consoloni L,Chiaretti S,Grassi G World J Gastroenterol. 2014 Jan 21;20(3):795-803 |
| Pubmed id:23967134 |
Specific inhibition of the redox activity of ape1/ref-1 by e3330 blocks tnf-α-induced activation of IL-8 production in liver cancer cell lines. Cesaratto L,Codarin E,Vascotto C,Leonardi A,Kelley MR,Tiribelli C,Tell G PLoS One. 2013;8(8):e70909 |
| Pubmed id:24116172 |
The expression of CD90/Thy-1 in hepatocellular carcinoma: an in vivo and in vitro study. Sukowati CH,Anfuso B,Torre G,Francalanci P,Crocè LS,Tiribelli C PLoS One. 2013;8(10):e76830 |
| Pubmed id:22309595 |
Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. Miura S,Mitsuhashi N,Shimizu H,Kimura F,Yoshidome H,Otsuka M,Kato A,Shida T,Okamura D,Miyazaki M BMC Cancer. 2012 Feb 6;12():56 |
| Pubmed id:21831731 |
Effects of E2F1-cyclin E1-E2 circuit down regulation in hepatocellular carcinoma cells. Farra R,Dapas B,Pozzato G,Scaggiante B,Agostini F,Zennaro C,Grassi M,Rosso N,Giansante C,Fiotti N,Grassi G Dig Liver Dis. 2011 Dec;43(12):1006-14 |
| Pubmed id:21882038 |
Mitochondrial bioenergetic profile and responses to metabolic inhibition in human hepatocarcinoma cell lines with distinct differentiation characteristics. Domenis R,Comelli M,Bisetto E,Mavelli I J Bioenerg Biomembr. 2011 Oct;43(5):493-505 |
| Pubmed id:20144681 |
Serum response factor depletion affects the proliferation of the hepatocellular carcinoma cells HepG2 and JHH6. Farra R,Dapas B,Pozzato G,Giansante C,Heidenreich O,Uxa L,Zennaro C,Guarnieri G,Grassi G Biochimie. 2010 May;92(5):455-63 |
| Pubmed id:20200618 |
Galectin-1 and its involvement in hepatocellular carcinoma aggressiveness. Spano D,Russo R,Di Maso V,Rosso N,Terracciano LM,Roncalli M,Tornillo L,Capasso M,Tiribelli C,Iolascon A Mol Med. 2010 Mar;16(3-4):102-15 |
| Pubmed id:20044606 |
Infrequent amplification of JUN in hepatocellular carcinoma. Endo M,Yasui K,Nakajima T,Gen Y,Tsuji K,Dohi O,Zen K,Mitsuyoshi H,Minami M,Itoh Y,Taniwaki M,Tanaka S,Arii S,Okanoue T,Yoshikawa T Anticancer Res. 2009 Dec;29(12):4989-94 |
| Pubmed id:19041685 |
Bortezomib arrests the proliferation of hepatocellular carcinoma cells HepG2 and JHH6 by differentially affecting E2F1, p21 and p27 levels. Baiz D,Pozzato G,Dapas B,Farra R,Scaggiante B,Grassi M,Uxa L,Giansante C,Zennaro C,Guarnieri G,Grassi G Biochimie. 2009 Mar;91(3):373-82 |
| Pubmed id:19142034 |
Contribution of AP-1 interference induced by TAC-101 to tumor growth suppression in a hepatocellular carcinoma model. Eshima K,Fukaya S,Sugimoto A,Mori T,Yokoi H,Yamamoto Y,Sugiura S,Honda S,Masuko N,Murakami K,Yamasaki Y,Kagechika H Tumour Biol. 2009;30(1):1-7 |
| Pubmed id:17825975 |
The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Grassi G,Scaggiante B,Farra R,Dapas B,Agostini F,Baiz D,Rosso N,Tiribelli C Biochimie. 2007 Dec;89(12):1544-52 |
| Pubmed id:15349899 |
Proteomic signature corresponding to alpha fetoprotein expression in liver cancer cells. Yokoo H,Kondo T,Fujii K,Yamada T,Todo S,Hirohashi S Hepatology. 2004 Sep;40(3):609-17 |
| Pubmed id:10766424 |
Involvement of insulin-like growth factor binding protein-3 in the retinoic acid receptor-alpha-mediated inhibition of hepatocellular carcinoma cell proliferation. Murakami K,Matsuura T,Hasumura S,Nagamori S,Yamada Y,Saiki I Cancer Lett. 2000 Apr 3;151(1):63-70 |
| Pubmed id:10933057 |
Invasiveness of hepatocellular carcinoma cell lines: contribution of membrane-type 1 matrix metalloproteinase. Murakami K,Sakukawa R,Ikeda T,Matsuura T,Hasumura S,Nagamori S,Yamada Y,Saiki I Neoplasia. 1999 Nov;1(5):424-30 |
| Pubmed id:9446792 |
Cyclin E overexpression responsible for growth of human hepatic tumors with p21WAF1/CIP1/SDI1. Tsuji T,Miyazaki M,Fushimi K,Mihara K,Inoue Y,Ohashi R,Ohtsubo M,Hamazaki K,Furusako S,Namba M Biochem Biophys Res Commun. 1998 Jan 14;242(2):317-21 |
| Pubmed id:8835345 |
Persistence of hepatitis C virus RNA in established human hepatocellular carcinoma cell lines. Tsuboi S,Nagamori S,Miyazaki M,Mihara K,Fukaya K,Teruya K,Kosaka T,Tsuji T,Namba M J Med Virol. 1996 Feb;48(2):133-40 |
| Pubmed id:1701409 |
Integration of hepatitis B virus DNA into cells of six established human hepatocellular carcinoma cell lines. Fujise K,Nagamori S,Hasumura S,Homma S,Sujino H,Matsuura T,Shimizu K,Niiya M,Kameda H,Fujita K Hepatogastroenterology. 1990 Oct;37(5):457-60 |
| Pubmed id:none |
樹立ヒト肝癌株細胞中のB型肝炎ウイルスゲノム 藤瀬 清隆, 藤多 和信, 永森 静志, 蓮村 哲, 本間 定, 筋野 甫, 松浦 知和, 清水 恵一郎, 新谷 稔, 大野 典也, 亀田 治男 肝臓 29(5), 697-698 (1988) |
| Pubmed id:none |
株化肝細胞癌の遺伝子発現 Gene expression in human hepatocellular carcinoma cell lines 村上 孝司 山田 雄次 済木 育夫 松浦 知和 蓮村 哲 永森 静志 組織培養研究 18(3), 221-228 (1999) |
| Images |
|---|
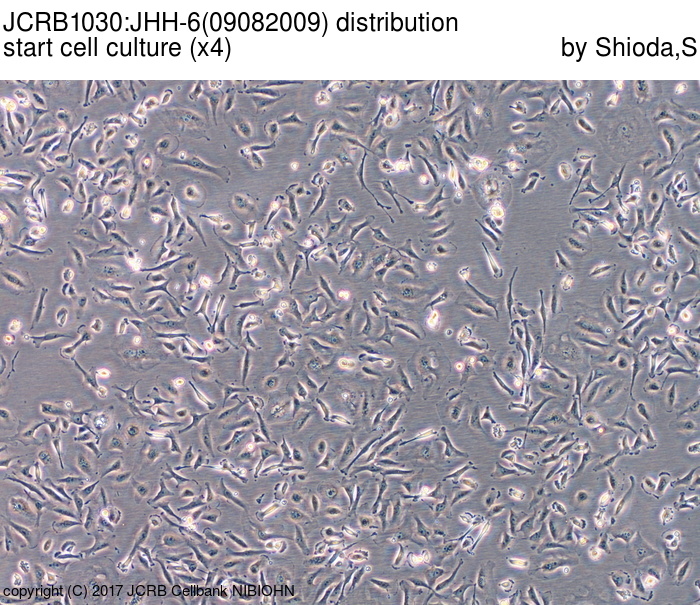 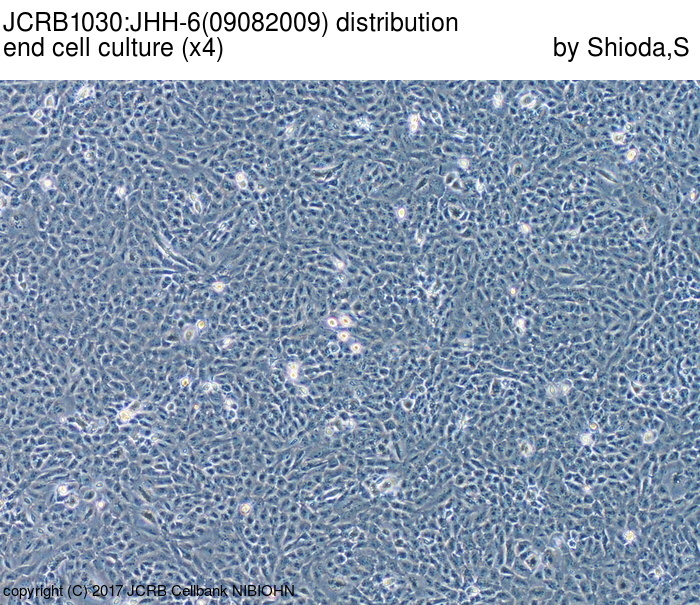 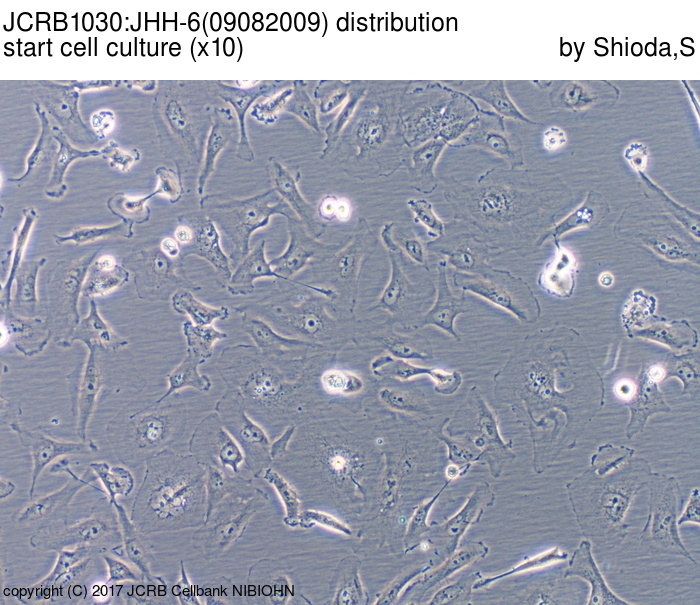 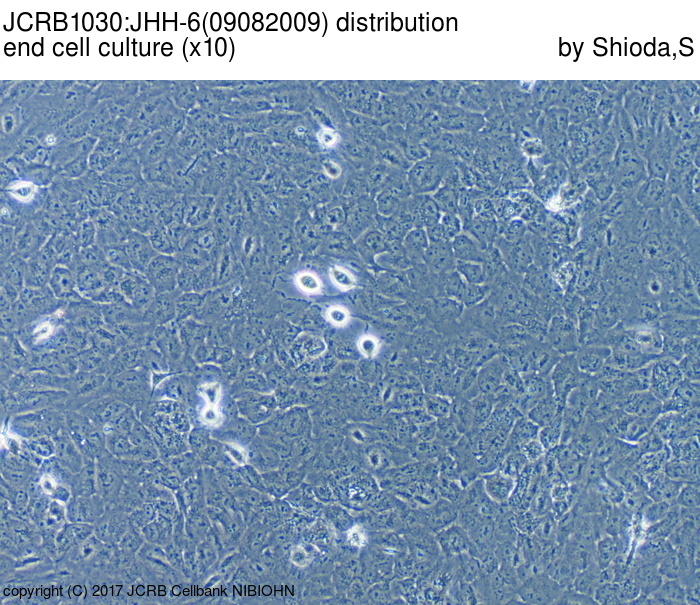 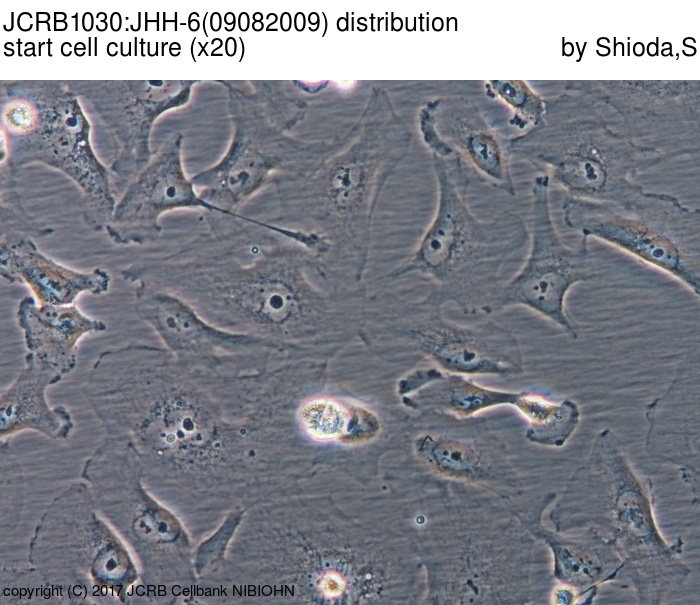 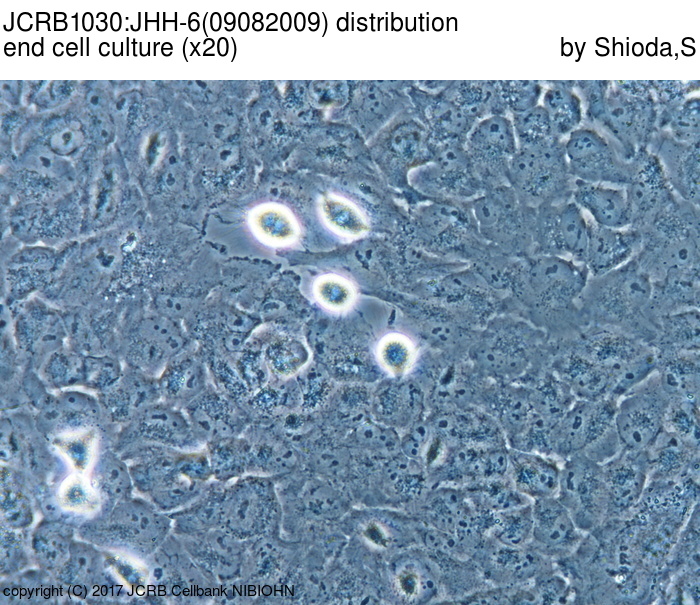   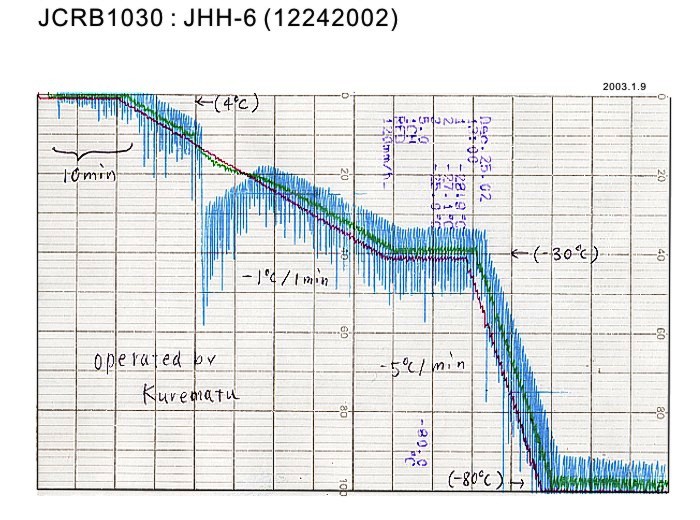 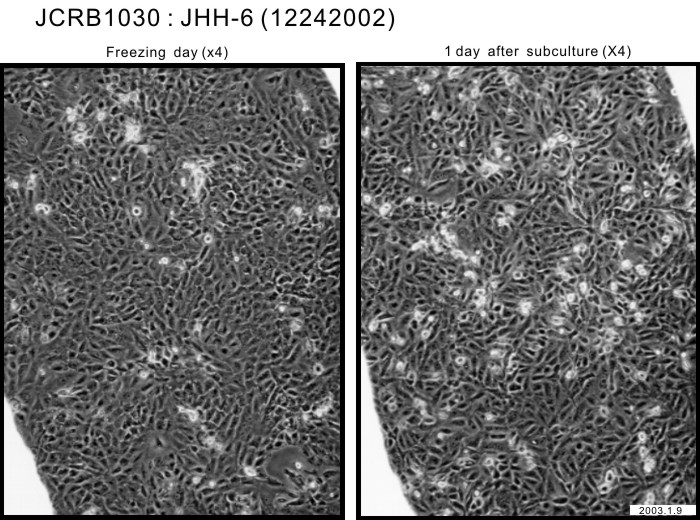 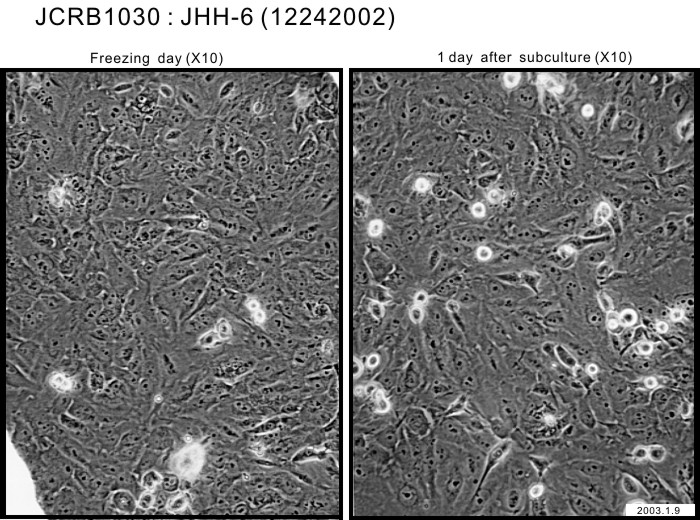 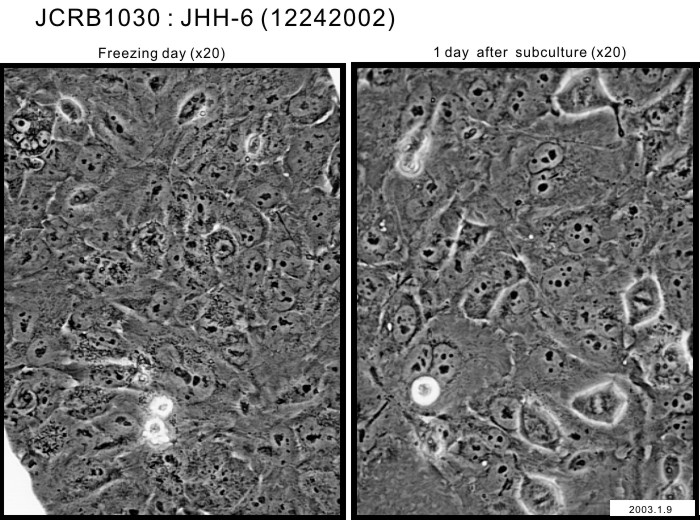 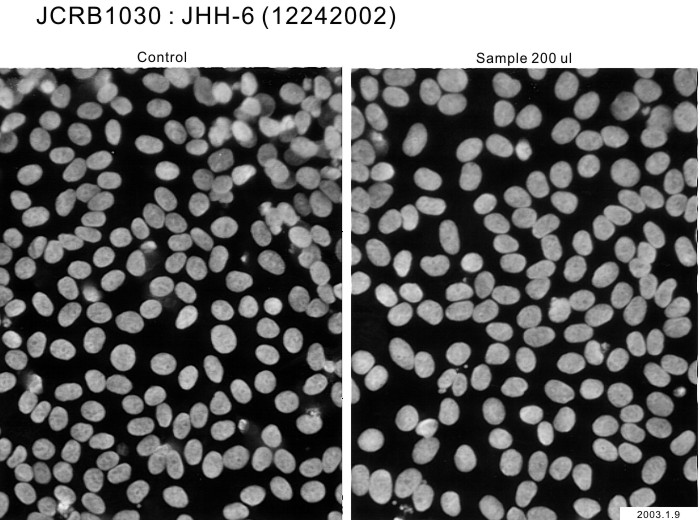  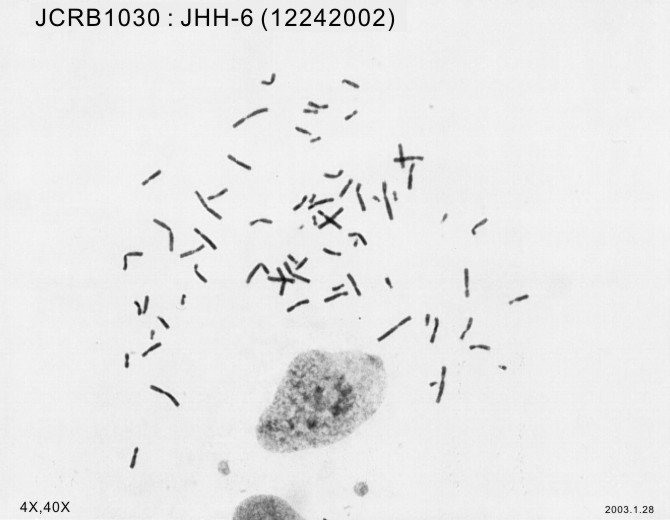 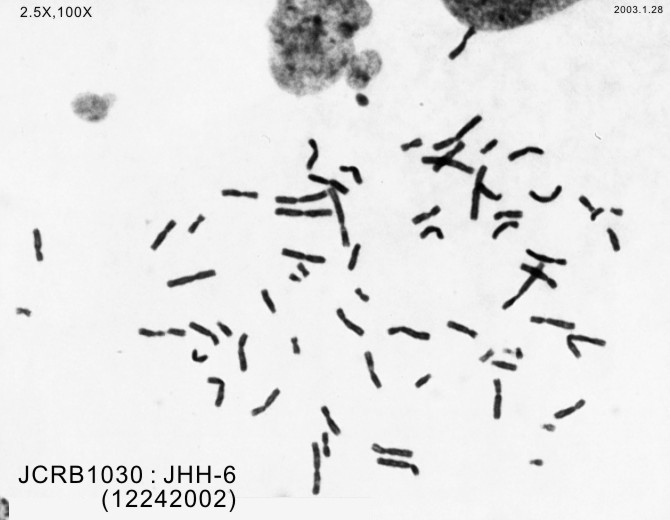 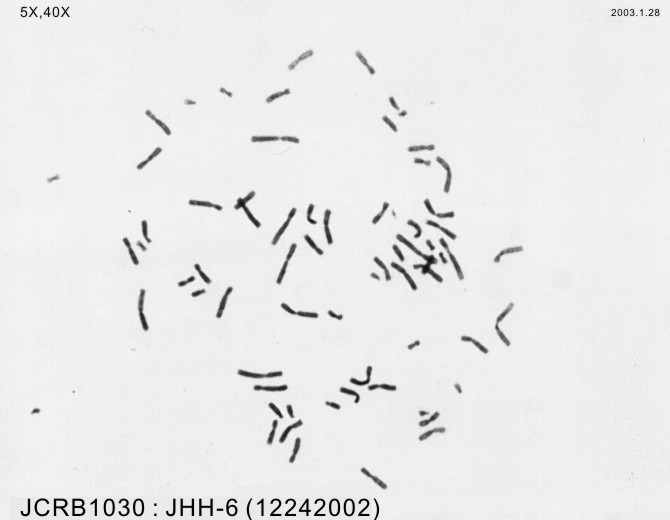     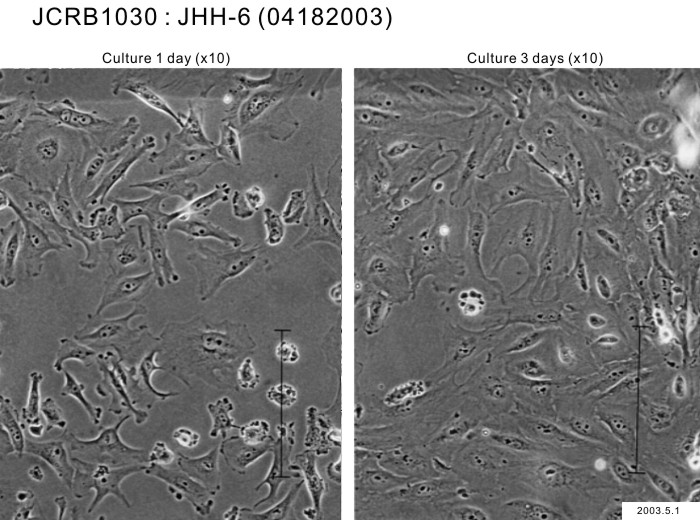 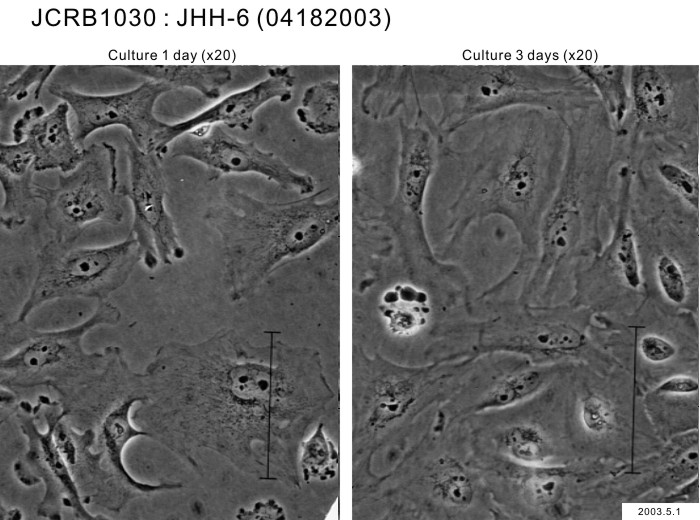 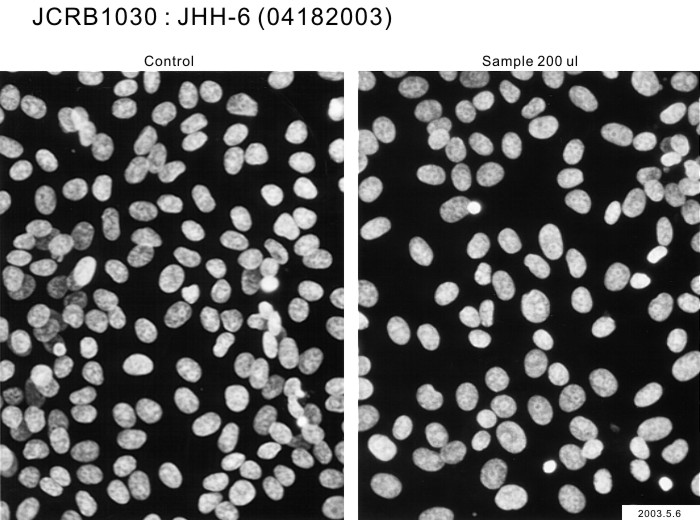 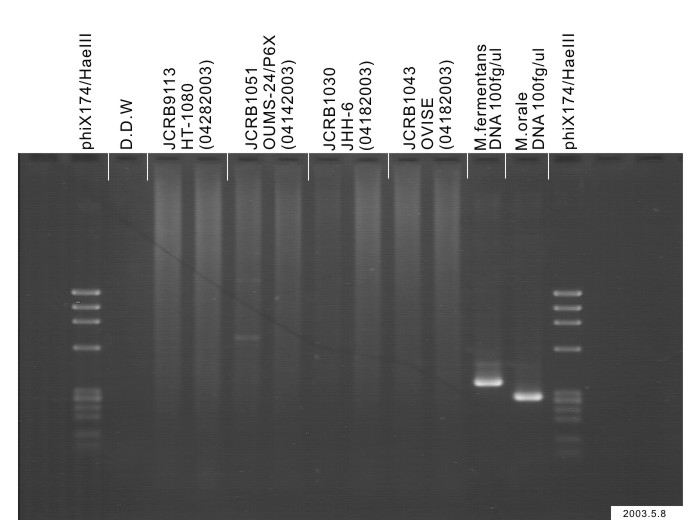 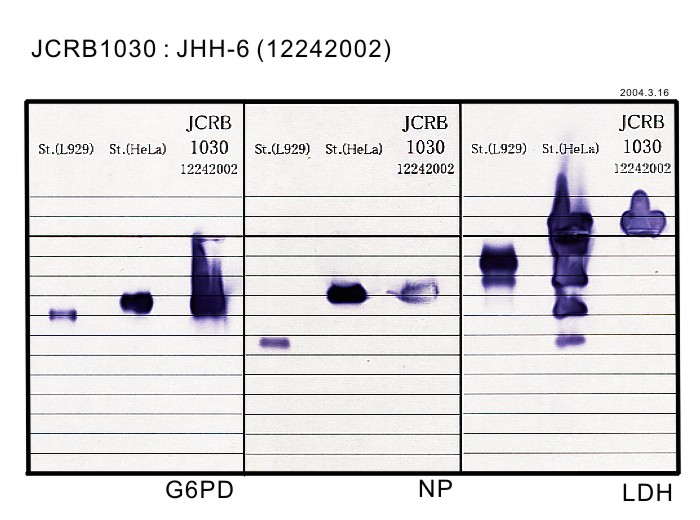 |
| Movies |
|---|



|
LOT Information
| Cell No. | JCRB1030 | Cell Name | JHH-6 |
|---|---|---|---|
| LOT No. | 09082009 | Lot Specification | distribution |
| Medium | Williamms' E medium with 10% fetal bovine serum (Intergen RB52305). | Temperature | 37 C |
| Cell Density at Seeding | 1.6x10^4 cells/sq.cm | Methods for Passages | Cells were harvested after treatment with 0.25%(GIBCO) trypsin and 0.02% EDTA. |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 2.0x10^6 |
| Viability at cell freezing (%) | 98.8 | Antibiotics Used | NT |
| Passage Number | p9* | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
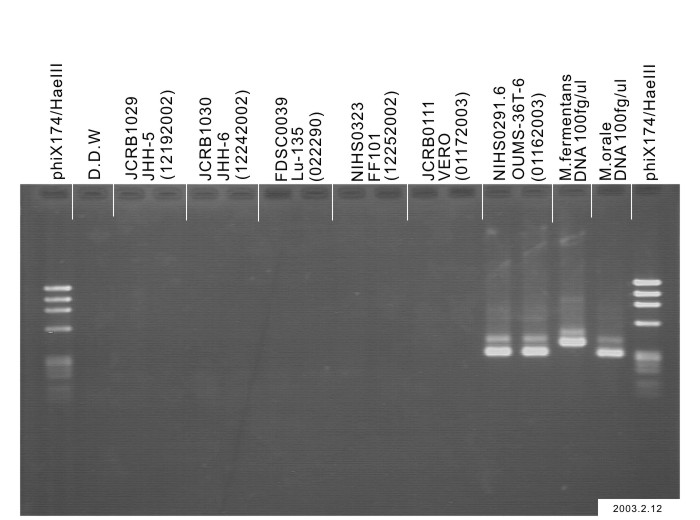
| Sterility: FUNGI | - | Isozyme Analysis | NT |
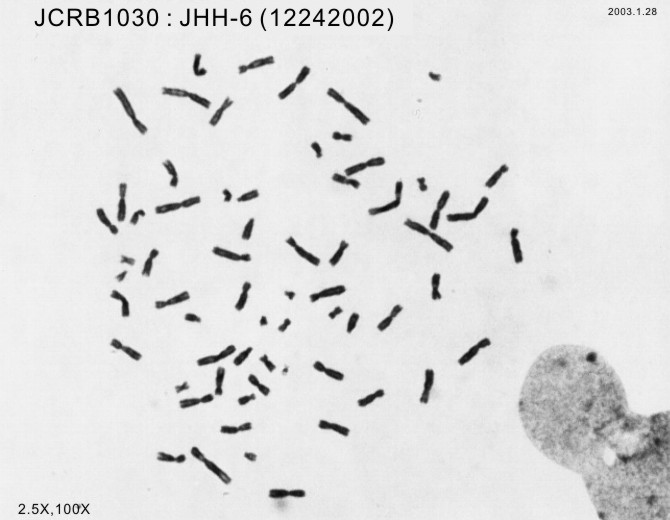
| Chromosome Mode | NT | Chromosome Information | NT |
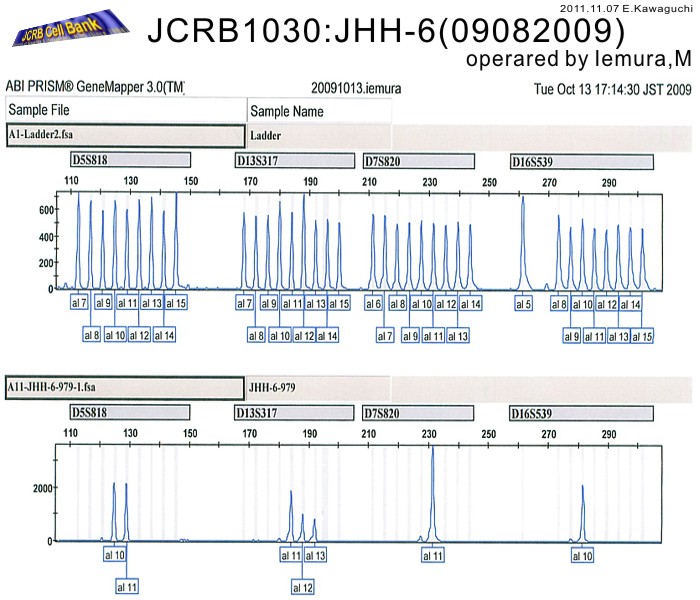
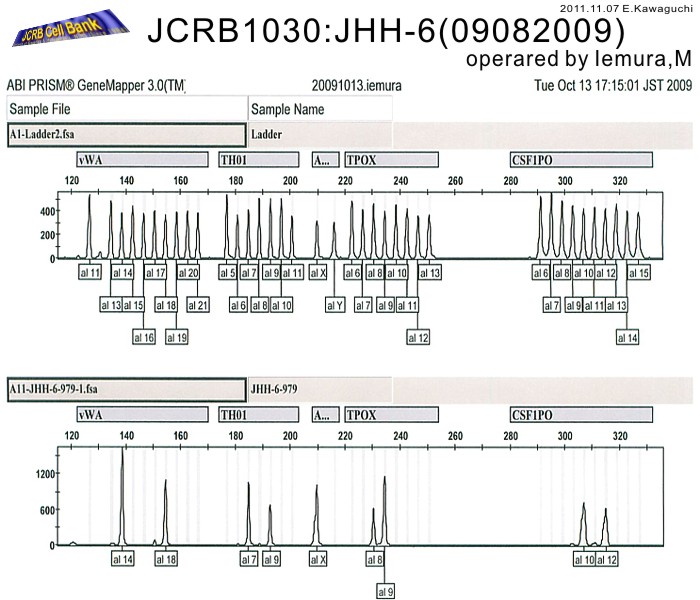
| Surface Antigen | NT | DNA Profile (STR) | D5S818:10,11 D13S317:11,12,13 D7S820:11 D16S539:10 VWA:14,18 TH01:7,9 AM:X TPOX:8,9 CSF1PO:10,12 |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | Cell Banker BLC-1(Nihon Zenyaku Industries) | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
        |
| Cell No. | JCRB1030 | Cell Name | JHH-6 |
|---|---|---|---|
| LOT No. | 04182003 | Lot Specification | distribution |
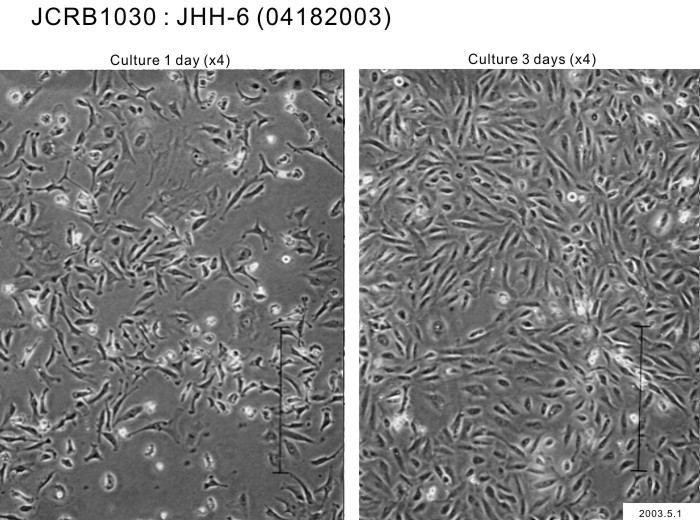
| Medium | William's E medium with 10% fetal calf serum (Intergen RB52305). | Temperature | 37 C |
| Cell Density at Seeding | 1.0 x 10^4 cells/sq.cm. | Methods for Passages | Cells harvested after treatment with 0.2% trypsin. Medium change twice a week. Subculture every 4-5 days. |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 1.1 x 10^6 |
| Viability at cell freezing (%) | 98.3 | Antibiotics Used | free |
| Passage Number | P8* | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | G6PD(type B), NP, LDH examined. Human not HeLa. |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | culture medium containing 5% DMSO. | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
      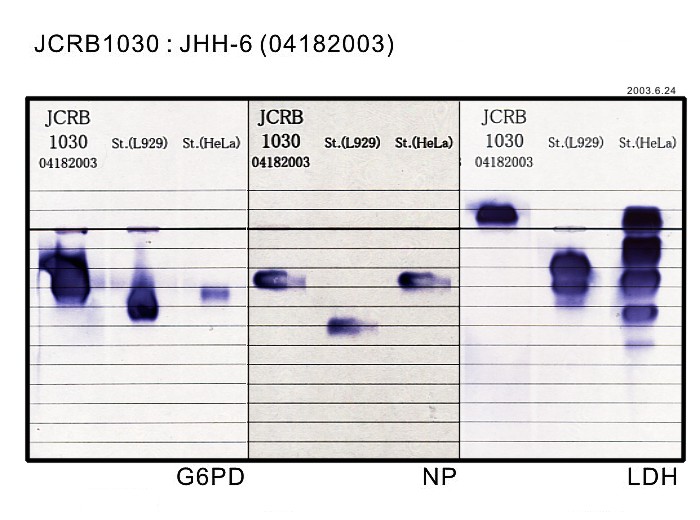 |
| Cell No. | JCRB1030 | Cell Name | |
|---|---|---|---|
| LOT No. | 03282003 | Lot Specification | distribution |
| Medium | William's E medium with 10% fetal bovine serum (FBS lot; BioWhittaker 0S069F) | Temperature | 37 C |
| Cell Density at Seeding | 2-3 x 10^4 cells/ml | Methods for Passages | 0.25% trypsin and 0.02% EDAT |
| Doubling Time | D.T. = ca. 30 hrs | Cell Number in Vial (cells/1ml) | 9.6 x 10^5 |
| Viability at cell freezing (%) | 86.4 | Antibiotics Used | free |
| Passage Number | Unknown (9 at bank) | PDL | |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | Confirmed as human by NP, G6PD (type B), LD. |
| Chromosome Mode | Chromosome Information | ||
| Surface Antigen | DNA Profile (STR) | ||
| Adhesion | Yes | Exoteric Gene | |
| Medium for Freezing | 10% DMSO, 20% FCS-culture | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | Additional information |
| Cell No. | JCRB1030 | Cell Name | JHH-6 |
|---|---|---|---|
| LOT No. | 05182017 | Lot Specification | distribution |
| Medium | Williams'E medium with 10% fetal bovine serum (FBS; Sigma Cat. # 172012) | Temperature | 37 C |
| Cell Density at Seeding | 1.2.6 - 9.3 x 10^4 cells/ml | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Doubling Time | approx. 21 hrs. | Cell Number in Vial (cells/1ml) | 1.9 x 10^6 |
| Viability at cell freezing (%) | 95.9 | Antibiotics Used | free |
| Passage Number | Unknown (9 at bank) | PDL | |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | |
| Chromosome Mode | Chromosome Information | ||
| Surface Antigen | DNA Profile (STR) | ||
| Adhesion | Yes | Exoteric Gene | |
| Medium for Freezing | 10% DMSO, 20% FBS - Williams'E medium | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | 95.9 | Additional information |
| Cell No. | JCRB1030 | Cell Name | JHH-6 |
|---|---|---|---|
| LOT No. | 06212018 | Lot Specification | distribution |
| Medium | Williams'E medium with 10% fetal bovine serum (FBS; Sigma Cat. # 172012) | Temperature | 37 C |
| Cell Density at Seeding | 2.0 - 3.4 x 10^4 cells/mL | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Doubling Time | approx. 29 hrs. | Cell Number in Vial (cells/1ml) | 8.7 x 10^5 |
| Viability at cell freezing (%) | Antibiotics Used | free | |
| Passage Number | Unknown (9 at bank) | PDL | |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | |
| Chromosome Mode | Chromosome Information | ||
| Surface Antigen | DNA Profile (STR) | ||
| Adhesion | Yes | Exoteric Gene | |
| Medium for Freezing | 10% DMSO, 20% FBS - Williams'E medium | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | 95.5 | Additional information |