JCRB1555 HAdpc-26-E6-Bmi-1-TERT
Cell information
Important Notice(s)Cell type:genetically-modified cells (View Pricing Information)
| JCRB No. | JCRB1555 | Cell Name | HAdpc-26-E6-Bmi-1-TERT |
|---|---|---|---|
| Profile | Immortalized human adipose tissue-derived mesenchymal stem cells. | Other Name | |
| Animal | human | Strain | |
| Genus | Homo | Species | sapiens |
| Sex | Age | ||
| Identity | available | Tissue for Primary Cancer | |
| Case history | Metastasis | No | |
| Tissue Metastasized | Genetics | Human adipose tissue-derived mesenchymal stem cells which were infected with recombinant retroviruses expressing the E6, Bmi-1, and hTERT. | |
| Life Span | infinite | Crisis PDL | |
| Morphology | fibroblast-like | Character | Derived from subcutaneous adipose tissue around mammary gland. hTERT, HPV E6 and bmi-1 genes were transfected. |
| Classify | transformed | Established by | Umezawa, A. |
| Registered by | Umezawa, A. | Regulation for Distribution | |
| Comment | Year | 2011 | |
| Medium | Poweredby 10 | Methods for Passages | When the cultures reached subconfluence, the cells were harvested with 0.25% trypsin and 1mM EDTA and replated with one-half of harvested cells. |
| Cell Number on Passage | Race | ||
| CO2 Conc. | 5% | Tissue Sampling | |
| Tissue Type | adipocyte |
| Detection of virus genome fragment by Real-time PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Detected DNA Virus | tested | Detected RNA Virus | tested | ||||||
| CMV | - | parvoB19 | - | HCV | - | HTLV-1 | - | ||
| EBV | - | HBV | - | HIV-1 | - | HTLV-2 | - | ||
| HHV6 | - | HTLV-1 | - | HIV-2 | - | HAV | - | ||
| HHV7 | - | HTLV-2 | - |
-/negative. +/positive. nt/not tested. (positive (+) does not immediately mean the production of infectious viral particles.) |
|||||
| BKV | - | HIV-1 | - | ||||||
| JCV | - | HIV-2 | - | ||||||
| ADV | - | HPV18 | - | ||||||
| Notes | |||||||||
| Images |
|---|
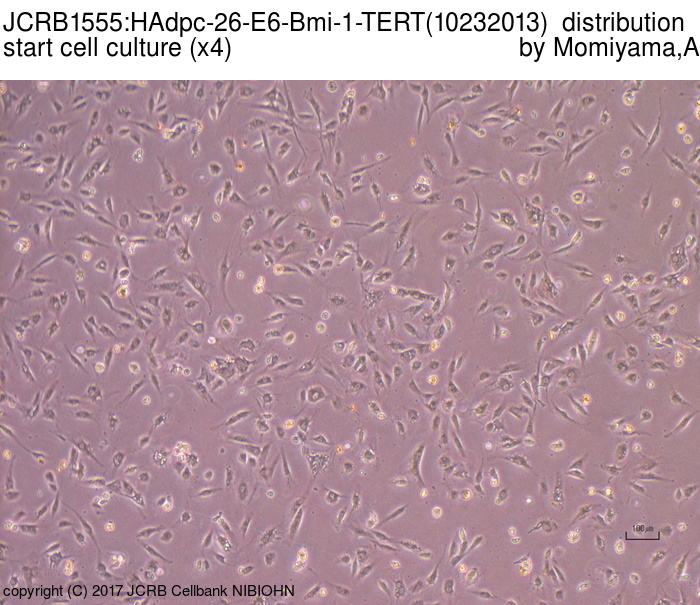 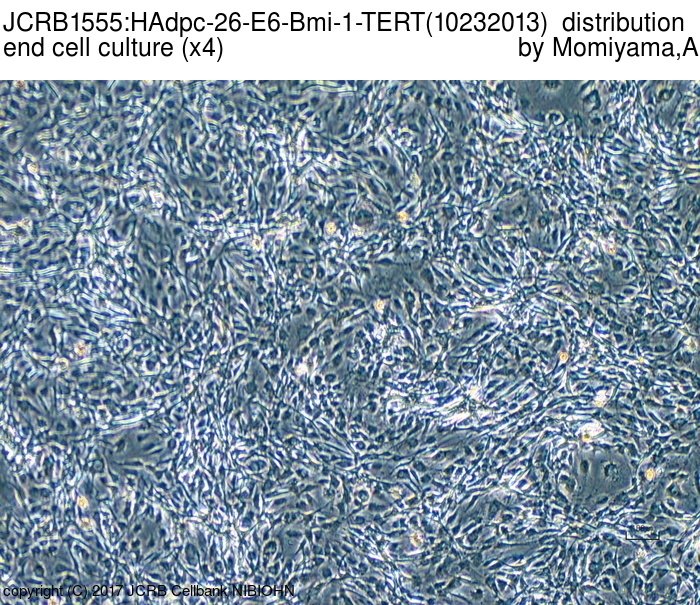 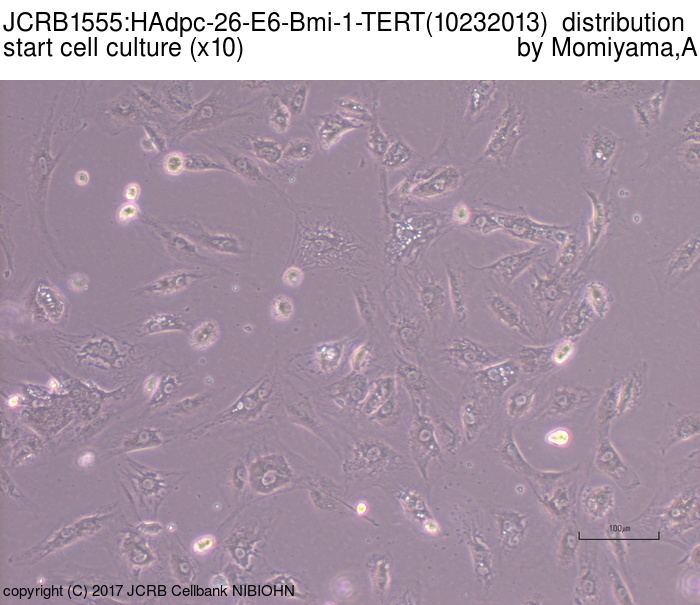 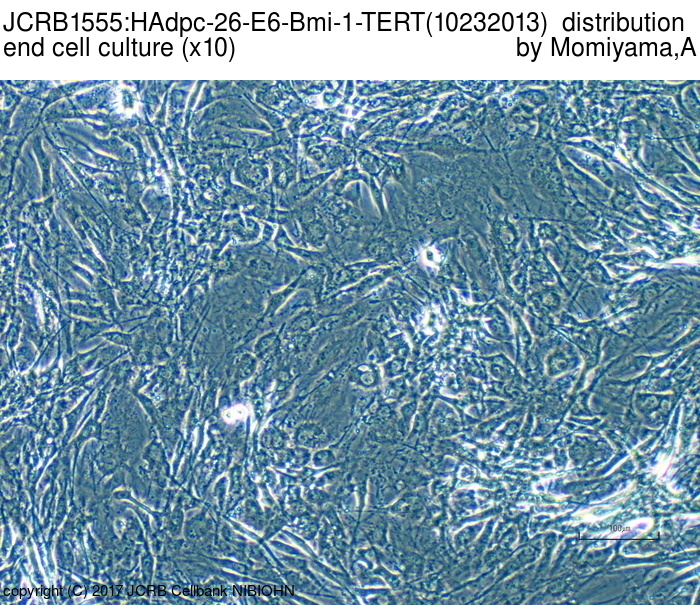 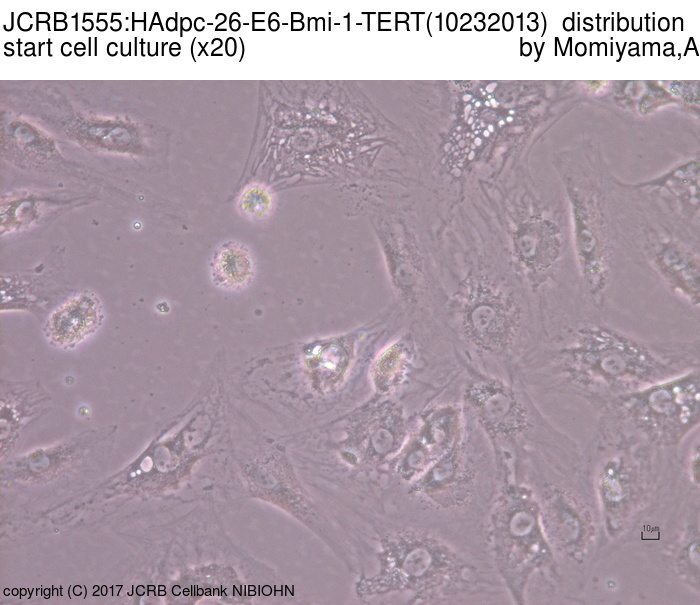 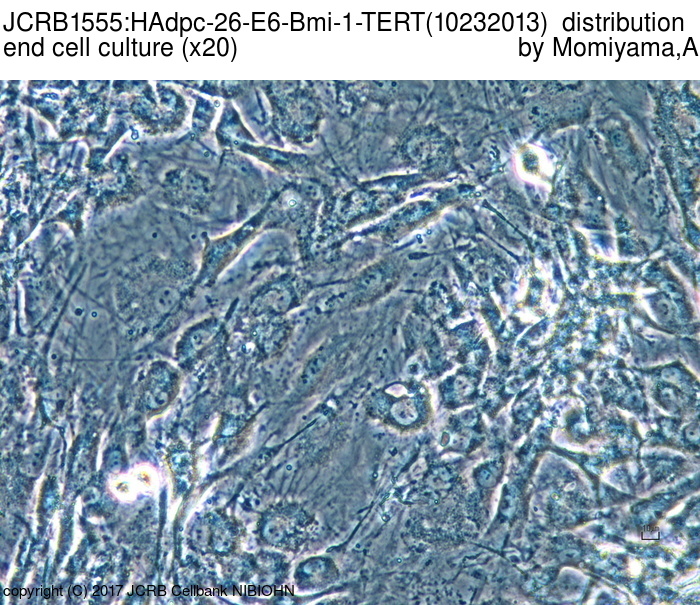 |
LOT Information
| Cell No. | JCRB1555 | Cell Name | HAdpc-26-E6-Bmi-1-TERT |
|---|---|---|---|
| LOT No. | 10232013 | Lot Specification | distribution |
| Medium | POWEREDBY10 | Temperature | 37 C |
| Cell Density at Seeding | 0.25-1.0x10^5 cells/sq.cm | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 1mM EDTA.(Split ratio=1/2-1/4) |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 4.39x10^6 |
| Viability at cell freezing (%) | 99.7 | Antibiotics Used | |
| Passage Number | p32 | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | NT |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | D5S818:12,13 D13S317:11 D7S820:11,12 D16S539:9,13 VWA:17,19 TH01:7 AM:X TPOX:8,11 CSF1PO:11,12 |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | Cell Banker BLC-1(Nihon Zenyaku Industries) | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
      |
| Cell No. | JCRB1555 | Cell Name | HAdpc-26-E6-Bmi-1-TERT |
|---|---|---|---|
| LOT No. | 01302023 | Lot Specification | distribution |
| Medium | Poweredby 10 medium | Temperature | 37C |
| Cell Density at Seeding | 2.0 - 9.0 x 10^4 cells/mL | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Doubling Time | approx. 34 hrs. | Cell Number in Vial (cells/1ml) | 1.0 x 10^6 |
| Viability at cell freezing (%) | 99 | Antibiotics Used | |
| Passage Number | P35 | PDL | |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | |
| Chromosome Mode | Chromosome Information | ||
| Surface Antigen | DNA Profile (STR) | D5S818:12,13 D13S317:11 D7S820:11,12 D16S539:9,13 VWA:17,19 TH01:7 AM:X TPOX:8,11 CSF1PO:11,12 |
|
| Adhesion | Yes | Exoteric Gene | |
| Medium for Freezing | Cell Banker BLC-1 (Nihon Zenyaku Industries) | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | 95 | Additional information |