JCRB1083 KYSE180
Cell information
Important Notice(s)On the MTA of KYSE series cell lines with Kyoto University (for profit organizations)
Announcement of unavailability of KYSE series cell lines to oversea users outside of Japan
Cell type:general cells (View Pricing Information)
| JCRB No. | JCRB1083 | Cell Name | KYSE180 |
|---|---|---|---|
| Profile | Human well differentiated squamous cell carcinoma from esophageal cancer. | Other Name | |
| Animal | human | Strain | |
| Genus | Homo | Species | sapiens |
| Sex | M | Age | 53 |
| Identity | available | Tissue for Primary Cancer | esophagus |
| Case history | esophagus cancer, T4N1Mo Stage 3, squamous cell carcinoma (SCC) | Metastasis | No |
| Tissue Metastasized | Genetics | cyclin D1 amplification, p53 mutation | |
| Life Span | infinite | Crisis PDL | |
| Morphology | epithelial-like | Character | Xenotransplantation positive. |
| Classify | tumor | Established by | Shimada,Y. |
| Registered by | Shimada,Y. | Regulation for Distribution | Cite reference {5368}. Company users require MTA from the originator. See additional information. |
| Comment | Distribution restricted to Japan(domestic use only). | Year | 2003 |
| Medium | Ham's F12 medium + RPMI1640 medium (1 to 1 mix) with 2% fetal calf serum. | Methods for Passages | Cells harvested after treatment with trypsin and EDTA. |
| Cell Number on Passage | split ratio = 1/5 | Race | Japanese |
| CO2 Conc. | 5 % | Tissue Sampling | esophagus |
| Tissue Type | Well differenciated squamous cell carcinoma. |
| Detection of virus genome fragment by Real-time PCR | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Detected DNA Virus | tested | Detected RNA Virus | tested | ||||||
| CMV | - | parvoB19 | - | HCV | - | HTLV-1 | - | ||
| EBV | - | HBV | - | HIV-1 | - | HTLV-2 | - | ||
| HHV6 | - | HTLV-1 | - | HIV-2 | - | HAV | - | ||
| HHV7 | - | HTLV-2 | - |
-/negative. +/positive. nt/not tested. (positive (+) does not immediately mean the production of infectious viral particles.) |
|||||
| BKV | - | HIV-1 | - | ||||||
| JCV | - | HIV-2 | - | ||||||
| ADV | - | HPV18 | - | ||||||
| Notes | |||||||||
| Reference | |
|---|---|
| Pubmed id:11092977 | Nonrandom chromosomal imbalances in esophageal squamous cell carcinoma cell lines: possible involvement of the ATF3 and CENPF genes in the 1q32 amplicon. Pimkhaokham A,Shimada Y,Fukuda Y,Kurihara N,Imoto I,Yang ZQ,Imamura M,Nakamura Y,Amagasa T,Inazawa J Jpn J Cancer Res. 2000 Nov;91(11):1126-33 |
| Pubmed id:10446988 | Lymph node metastasis is associated with allelic loss on chromosome 13q12-13 in esophageal squamous cell carcinoma. Harada H,Tanaka H,Shimada Y,Shinoda M,Imamura M,Ishizaki K Cancer Res. 1999 Aug 1;59(15):3724-9 |
| Pubmed id:9699676 | Methylation of the 5' CpG island of the FHIT gene is closely associated with transcriptional inactivation in esophageal squamous cell carcinomas. Tanaka H,Shimada Y,Harada H,Shinoda M,Hatooka S,Imamura M,Ishizaki K Cancer Res. 1998 Aug 1;58(15):3429-34 |
| Pubmed id:9033652 | Multiple types of aberrations in the p16 (INK4a) and the p15(INK4b) genes in 30 esophageal squamous-cell-carcinoma cell lines. Tanaka H,Shimada Y,Imamura M,Shibagaki I,Ishizaki K Int J Cancer. 1997 Feb 7;70(4):437-42 |
| Pubmed id:8575860 | Characterization of p53 gene mutations in esophageal squamous cell carcinoma cell lines: increased frequency and different spectrum of mutations from primary tumors. Tanaka H,Shibagaki I,Shimada Y,Wagata T,Imamura M,Ishizaki K Int J Cancer. 1996 Jan 26;65(3):372-6 |
| Pubmed id:9816044 | p53 mutation, murine double minute 2 amplification, and human papillomavirus infection are frequently involved but not associated with each other in esophageal squamous cell carcinoma. Shibagaki I,Tanaka H,Shimada Y,Wagata T,Ikenaga M,Imamura M,Ishizaki K Clin Cancer Res. 1995 Jul;1(7):769-73 |
| Pubmed id:7913084 | Analysis of gene amplification and overexpression in human esophageal-carcinoma cell lines. Kanda Y,Nishiyama Y,Shimada Y,Imamura M,Nomura H,Hiai H,Fukumoto M Int J Cancer. 1994 Jul 15;58(2):291-7 |
| Pubmed id:8518899 | Prognostic significance of cell culture in carcinoma of the oesophagus. Shimada Y,Imamura M Br J Surg. 1993 May;80(5):605-7 |
| Pubmed id:1728357 | Characterization of 21 newly established esophageal cancer cell lines. Shimada Y,Imamura M,Wagata T,Yamaguchi N,Tobe T Cancer. 1992 Jan 15;69(2):277-84 |
| Images |
|---|
         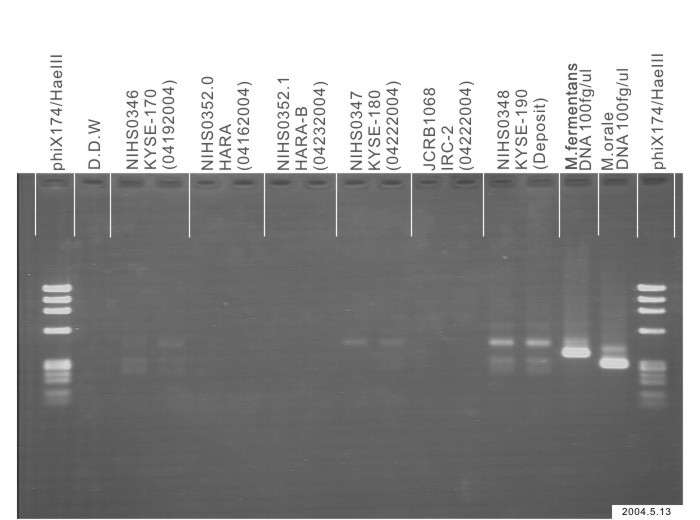 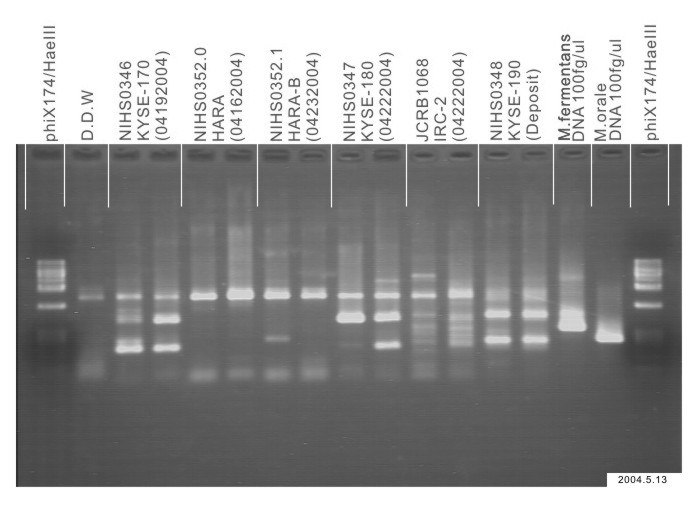     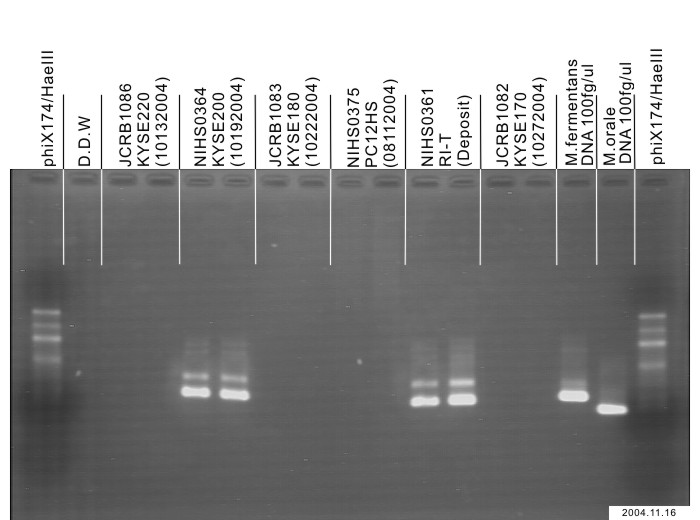         |
LOT Information
| Cell No. | JCRB1083 | Cell Name | KYSE180 |
|---|---|---|---|
| LOT No. | 07252008 | Lot Specification | distribution |
| Medium | RPMI1640(GIBCO) and Ham's F12 medium(GIBCO)(1/1) with 2% heat inactivated fetal bovine serum(Hyclone GRD0051) | Temperature | 37 C |
| Cell Density at Seeding | 1.9x10^4 cells/sq.cm | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 2.2x10^6 |
| Viability at cell freezing (%) | 98.9 | Antibiotics Used | free (this lot was pretreated with plasmocin) |
| Passage Number | p715 | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | NT |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | D5S818:8,11 D13S317:9,12 D7S820:9,12 D16S539:9,10 VWA:16,17 TH01:7 AM:X TPOX:11 CSF1PO:12,13 |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | Cell Banker BLC-1(Nihon Zenyaku Industries) | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
         |
| Cell No. | JCRB1083 | Cell Name | KYSE180 |
|---|---|---|---|
| LOT No. | 08012008 | Lot Specification | distribution |
| Medium | RPMI1640(GIBCO) and Ham's F12 medium(GIBCO)(1/1) with 2% heat inactivated fetal bovine serum(Hyclone GRD0051) | Temperature | 37 C |
| Cell Density at Seeding | 1.9x10^4 cells/sq.cm | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin and 0.02% EDTA. |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 4.0x10^6 |
| Viability at cell freezing (%) | 99.0 | Antibiotics Used | free (this lot was pretreated with BM cyclin 1and |
| Passage Number | p716 | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | NT |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | D5S818:8,11 D13S317:9,12 D7S820:9,12 D16S539:9,10 VWA:16,17 TH01:7 AM:X TPOX:11 CSF1PO:12,13 |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | Cell Banker BLC-1(Nihon Zenyaku Industries) | CO2 Conc. | 5 % |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
         |
| Cell No. | JCRB1083 | Cell Name | KYSE180 |
|---|---|---|---|
| LOT No. | 10222004 | Lot Specification | distribution |
| Medium | 1 to 1 mix of Ham's F12 and RPMI1640 medium containing 2% fetal calf serum. | Temperature | 37 C |
| Cell Density at Seeding | 4.0 x 10^4 cells / sq. cm | Methods for Passages | Cells were harvested after treatment with 0.25% trypsin about 5 min. Subculture once a week and medium change every 2-4 days. |
| Doubling Time | NT | Cell Number in Vial (cells/1ml) | 3.8x10^6 |
| Viability at cell freezing (%) | 96 | Antibiotics Used | free |
| Passage Number | P715 | PDL | NT |
| Sterility: MYCOPLASMA | - | Sterility: BACTERIA | - |
| Sterility: FUNGI | - | Isozyme Analysis | NT |
| Chromosome Mode | NT | Chromosome Information | NT |
| Surface Antigen | NT | DNA Profile (STR) | |
| Adhesion | Yes | Exoteric Gene | NT |
| Medium for Freezing | Culture medium containing 5% DMSO at final concentration. | CO2 Conc. | 5% |
| Viability immediately after thawing (%) | Additional information |
| Images |
|---|
         |